NERLSCD/MADSSCi 2019 Detailed Program
NERLSCD MADSSCi 19
Exploring New Technologies to Drive Advances in Basic, Clinical and Translational Research
November 6-8, 2019 – Philadelphia PA
Detailed Program
Pre-Conference Work Shops at Hotel 201
Cytek
Meet with Aurora users and the Cytek R&D team in a casual environment to consult on getting the most out of your Aurora. Cytek’s team will also present Aurora Best Practices.”
Time: 9:00-4:00
Location: Ballroom D
Agilent
Come meet our experts and learn how to maximize the potential of your core facility. We’ll show you how to deploy a robust, flexible core facility management system.
- Genomics – 1:30 – 3:00 pm
- Coffee and Refreshments Break 2:40 – 3:00 pm
- iLab – 3:00 – 5:00 pm
Time: 1:30-5:00
Location: Salon Rooms 5&6
New England Biolabs (NEB)
Join NEB on Tuesday, November 6th from 2:30-3:45pm to celebrate the 10th anniversary of the NEBNext product portfolio.
Time: 2:30-3:45
Location: Salon Rooms 3&4
NYCAN Core Administrators Pre-Conference Meeting
Come join us for our annual Regional Core Administration Meeting. This is a forum, open to all, not just those from the New York area where we all get to exchange ideas and propose varied approaches to Cores and career-related problem-solving. Walk-in at any time during this session. Light refreshments will be provided.
Time: 4:30-5:30
Location: Hotel 201 – Terrace Lounge
Illumina
Pre-Conference Get-together
Join Illumina for an informal round-table discussion in support of our NGS platforms. Representatives from sales, field applications and field service will be available to answer questions and address concerns.
Time: 5:00-6:00
Location: Salon Rooms 3&4
Opening Social at Franklin Institute
The Franklin Institute, a science museum and the center of science education and research in Philadelphia, Pennsylvania, is named after the American scientist and statesman Benjamin Franklin and houses the Benjamin Franklin National Memorial. Founded in 1824, the Franklin Institute is one of the oldest centers of science education and development in the United States.
Explore open exhibits at the institute including The Brain, Electricity, and Changing Earth
Enjoy drinks and small plates
Opening Remarks: Andrew Chitty, PhD, Oregon Health and Science University, ABRF President
Details: Learn about the current status of our parent organization, the Association of Biomolecular Resource Facilities, including updates, on-going initiatives, and more!
Time: 6:15-9:00
Location: Franklin Institute
Directions from Hotel 201: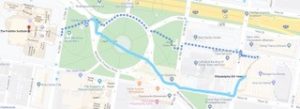
Opening Remarks:
Louis J. Soslowsky, PhD
Fairhill Professor Orthopaedic Surgery
Associate Dean for Research Integration,
Perelman School of Medicine,
University of Pennsylvania
Time: 8:30 am
Location: Liberty Ballroom D
Keynote
- Monica Guzman, PhD
Associate Professor of Pharmacology in Medicine
Weill Cornell Medical College
Title: Tailoring therapeutic approaches for selective targeting of leukemia stem cells
Abstract
In efforts to improve AML therapy, we have been searching for strategies that allow the effective elimination of LSCs. We and others, have demonstrated that LSCs possess unique molecular features that distinguish them from their normal hematopoietic stem cells (HSCs) and that such can be exploited to selectively eliminate LSCs without harming HSCs. However, as there heterogeneity in AML, it is important to understand the cellular dependencies of AML cells to be able to tailor therapies to individual patients.
Speaker Bio
Dr. Guzman is an Associate Professor of Pharmacology in Medicine, appointed in the Division of Hematology & Medical Oncology in the Department of Medicine. She was recruited to Weill Cornell Medical College in 2009 from the University of Rochester. As the first scientist to identify dysregulated gene networks in leukemia stem cells and capitalize on this dysregulation to develop therapies to destroy them, she is widely viewed as one of the world’s foremost experts in therapeutic targeting of leukemia stem cells. She has authored or co-authored numerous papers in the field and has received funding from the NIH, Leukemia & Lymphoma Society, V Foundation, and other organizations. Her passion is to perform world class translational research through close interactions between research and clinic to accelerate the development of new and more effective leukemia treatments that ablate leukemia stem cells.
Location: Liberty Ballroom
Time: 8:40 am
Platinum Plus Vendor Sponsor Plenary – Cytek
Speaker
Chris Fleming, PhD
Technical Application Specialist
Cytek Biosciences
Title: An Introduction to Full Spectrum Flow Cytometry
Abstract
Full spectrum flow cytometry enables the achievement of high quality, high complexity 40 color immunophenotyping panel from a single sample tube. Join us to learn more about the Cytek Aurora and the advantages provided by full spectrum cytometry.
Location: Liberty Ballroom D
Time: 9:40 am
Concurrent Sessions Block 1
Time: 10:15-11:30 am
Track – Genomics
Title: Emerging Techniques in Translational Genomics
Location: Freedom Ballroom G
Speakers
- Kevin Gerrish, PhD
Director, Molecular Genomics Core
National Institute of Environmental Health Sciences
- Samantha Sevilla Chill, M.S.
Development Associate III, Cancer Genomics Research Laboratory
National Cancer Institute
Abstract
This session will focus on two emerging technologies in translational genomics: circulating cell free DNA and microbiome analyses.
Track – Flow Cytometry
Title: Trending Topics in Flow Cytometry
Location: Freedom Ballroom F
Speakers:
- Kathleen Daniels, SCYM (ASCP)CM
Manager, Flow Cytometry Core Facility
ISAC SRL Emerging Leader
- Evan Jellison, PhD
Director, Flow Cytometry
University of Connecticut, Health
- Joshua Welsh, PhD
Visiting Postdoctoral Fellow
ISAC Marylou Ingram Scholar, Translational Nanobiology Section, Laboratory of Pathology
Center for Cancer Research
National Cancer Institute
Abstract
This session will focus on currently trending topics in flow cytometry including: Best practices involving Compensation/Reference controls (Beads vs. Cells); new software tool for scatter and fluorescence calibration for standardized reporting; and Tips for cell sorting when nucleic acids are the actual targets.
Speaker 1: Talk Title: “Beads vs. Cells: Compensating with (un)mixed feelings. Description: With the practice of multiparametric cytometry comes the need to correct for the spectral overlap of fluorochromes that are used in combination. Both beads and cellular single color controls can be used for this correction via unmixing or compensation, and here we will discuss an in depth comparison between these controls that are so often used in cytometry experiments.
Speaker 2: Talk Title: “When a genomics giant moves in next door…Tips for sorting when nucleic acids are the actual targets” Description: This talk will cover instrument setup, including 96-well and 384-well deposition, nozzle selection, sort buffer selection, collection buffer selection, sorting for 10x sequencing, sorting for other sequencing (including BD genomics offerings), sorting into dry wells, and sorting frozen biobanked tissue. This session is intended to be interactive with audience participation.
Speaker 3: Talk title: A light scatter and fluorescence calibration tool for reporting small particle data in standard units. Description: FCMPASS is a free software tool that can perform light scatter calibration, fluorescence calibration and write parameters and output reports in standard units for publication. The software also allows either diameter or refractive index approximation from light scatter signals.
Track – Bioinformatics
Title: Understanding Complexity in Single Cell Transcriptomics
Sponsor: Advaita Bioinformatics
Location: Freedom Ballroom E
Speakers
- Ilya Korsunsky, PhD
Department of Medicine, Brigham and Women’s Hospital
Department of Biomedical Informatics, Harvard Medical School
- Richard McEachin, PhD
Adviata Bioinformatics
- David Van Dijk, PhD
Yale University
Abstract
Summary:
The emerging diversity of single cell RNAseq datasets allows for the full transcriptional characterization of cell types across a wide variety of biological and clinical conditions. However, it is challenging to analyze them together, particularly when datasets are assayed with different technologies, because biological and technical differences are interspersed. We present Harmony (https://github.com/immunogenomics/harmony), an algorithm that projects cells into a shared embedding in which cells group by cell type rather than dataset-specific conditions. Harmony simultaneously accounts for multiple experimental and biological factors. In six analyses, we demonstrate the superior performance of Harmony to previously published algorithms while requiring fewer computational resources. Harmony enables the integration of ~106 cells on a personal computer. We apply Harmony to PBMCs from datasets with large experimental differences, 5 studies of pancreatic islet cells, mouse embryogenesis datasets, and cross-modality spatial integration
Speaker 2:
10 lessons learned from 10 years experience as bioinformatics core directors
You have a challenging task. You provide expert bioinformatics services to your organization. You have great analysts in your group, but not enough of them. Your organization does extremely important work in biomedical research, but you have to live on a budget. You do a range of analyses where you need to understand the biological relevance of a gene set (e.g. single-cell expression profile). Your customers are domain experts who love to dig into their results but don’t want to become bioinformaticists.
In this brief talk, we will discuss 10 lessons learned from 10 years experience directing bioinformatics cores at the University of Michigan and Wayne State University. Though these lessons come from bioinformatics cores, they apply equally to any data analysis core (e.g. biostatistics, genomics, proteomics). Learn how to get the best results to your customers, maximize the benefit to these domain experts, and maximize the positive impact of your core on your organization.”
Track – Administration
Title: Websites and Social Media: What works for your core facility?
Location: Freedom Ballroom H
Speakers:
- Lisa MacDowell, MBA
Children’s Hospital of Philadelphia
- Susanna Perkins, M.S.
University of Massachusetts
Abstract
- How does a decentralized institution manage a social media presence?
- What do you post? How do you get your information?
- Who does the regular updates?
- What type of feedback do you get to see if it’s working? Are your “followers” really individuals that will use your Cores or just friends of friends? How can you tell if you are attracting real paying customers?
- How do you fund this initiative?
- Do you use any sort of incentives to get people to follow you and retweet posts? If so, what are they and who funds them?
With Social Media Platforms continuing to be a robust form of communication beyond your Institution’s 4 walls, how do you keep up? Is your website really an advantage to your customer base or just a page of pricing?
This session will address these questions and more through interactive discussions with the entire audience. Please bring your questions, ideas, and success stories to share with others who are either just starting the process of on-line marketing or have a complete package of offerings (website, facebook, twitter, Instagram, linked-in). For those new to this forum, we hope the shared ideas will give you areas to focus on that will work for your Core(s); for those fully engaged, we hope that you can share what works for you and why and hope that you walk around with a new idea to implement
Concurrent Sessions Block 2: Technology 101s
Time: 1:00-2:00 pm
About: Always wanted to learn the basics of another technology but never had time? These 101 sessions are designed to introduce you to the foundations of a technology other than your own area of expertise.
Track – Genomics
Title: Genomics 101
Location: Freedom Ballroom G
Speakers:
- Roxann Ashworth, MHS
Laboratory Co-Director, Genetic Resources Core Facility DNA Services
Johns Hopkins University
- Deborah Hollingshead, MS
Assistant Director, Genomics Research Core
University of Pittsburgh
Abstract
This session is designed to explain to non-genomics people what happens in the wide variety of genomics core facilities at academic institutions.
Track – Flow Cytometry
Title: Flow Cytometry 101
Location: Freedom Ballroom F
Speakers
- Peter Lopez, PhD
New York University
- Julie Nelson, MS
Center for Tropical and Emerging Global Disease, Cytometry Shared Resource Laboratory
University of Georgia
Abstract
This session will survey the history and basis on the technology, the wide range of diverse applications, and the latest developments in the field of flow cytometry.
Track – Mass Spectrometry
Title: Mass Spectrometry 101
Location: Freedom Ballroom E
Speakers
- Jace Jones, PhD
Assistant Professor
Associate Director, Mass Spectrometry Center
Department of Pharmaceutical Sciences
University of Maryland School of Pharmacy
- Yan Wang, PhD
Director, Mass Spec Lab
National Institutes of Health
Abstract
This is a crash course on basics of mass spectrometry and its application in core facilities. We will start with a quick overview of different types of mass spectrometers used in core facilities, followed by applications of mass spectrometry in the world of small molecule analysis, (which covers mostly everything other than proteins). We will spend more time going through basics of proteomics and what is required to set up proteomics service. Both speakers are career mass spec core directors and will be available for questions after the talk.
Track – Imaging
Title: Imaging 101 – what we do and what is new in imaging cores
Location: Freedom Ballroom H
Speakers
- Elke Küster-Schöck
Platform for Imaging by Microscopy
CHU Sainte-Justine/Université de Montréal, Montréal
Québec, Canada
- Melina Jaramillo Garcia
Molecular and Cellular Microscopy Platform
Douglas Hospital Research Centre, McGill University
Quebec, Canada
- Andrea Stout, PhD
Cell and Developmental Biology Microscopy Core,
Perelman School of Medicine at the University of Pennsylvania
Abstract
Imaging plays an important role in the life sciences, and modern imaging technologies have evolved over the past years and decades. In this session, which is geared towards an audience from outside the field, we discuss the types of imaging typically found in the context of life science research. Focussing on light microscopy, we explore standard technologies and newer developments like super-resolution microscopy. In a final section, we highlight the structure of light microscopy core facilities and some of the administrative challenges that set imaging labs apart from cores that mostly process samples.
Concurrent Sessions Block 3
Time: 2:00-3:25 pm
Track – Genomics
Title: Genomic Technologies: Best Practices for Core Centers
Location: Freedom Ballroom G
Sponsor: Qiagen
Speaker:
- Jonathan Shaffer, PhD, MBA
Associate Director, NGS Assay Research & Development
- Jonathan Shaffer, PhD, MBA
Title: QIAseq FastSelect: One-Step, 14 Minute Removal of rRNA During Whole Transcriptome NGS Library Prep
Abstract
Whole transcriptome next-generation sequencing (NGS) enables the characterization of both mRNA and long noncoding RNAs (lncRNAs) from biological samples. For optimal results, significantly overrepresented RNAs, such as ribosomal RNA (rRNA) and globin mRNA must be removed. Elimination of these contaminating RNAs simultaneously increases sensitivity and decreases the sequencing budget required per sample.
To remedy the shortcomings associated with existing depletion methods (poor efficiency, distorted transcriptomic profiles, ill-suited for damaged samples, time consuming), we have developed QIAseq FastSelect –rRNA HMR and –Globin. The FastSelect technology utilizes a novel, one-step rRNA depletion method that prevents cDNA synthesis of unwanted RNAs in cell, tissue, and whole blood RNA, whether the molecules are intact or degraded. Furthermore, FastSelect is compatible with virtually all stranded RNAseq library methods, and works with human, mouse and rat samples, as well as other model organisms. Custom kits are also available for any species or RNA.
Session Speakers:
- Host: Bruce Kingham
Director, Sequencing & Genotyping Center
University of Delaware
- Jennifer Holbrook
Assistant Director Biomolecular Core Lab
Nemours Children’s Health System
- Christian H. Lytle
Administrative Coordinator for Shared Resources
Norris Cotton Cancer Center
Laboratory Manager
Molecular Biology Core Facility
Geisel School of Medicine at Dartmouth
- Peter Schweitzer, PhD
Director, Genomics Facility
Cornell University
- Scott Tighe
Technical Director
University of Vermont
Abstract:
Genomic technologies continue to expand at an unprecedented rate in core facilities, with user demands spanning from single cell transcriptomics to whole genome assemblies and full systems biology characterizations. There has never been greater need for revolutionary technologies that deliver such genome information. The demand for genome data has accelerated the development of next-generation genomic technologies. Considering the diversity of available features, it’s expected that multiple platforms will coexist in the same genomic space, with some having clear advantages over others for particular applications. The economy of scale associated with generating large genomic data sets using next generation technologies is the primary advantage over conventional methods. Costs will continue to decline in the foreseeable future, although the cost reduction should be evaluated against the quality of the data produced. All of these factors will necessitate additional scrutiny by core facilities and the end users.
In this session, we will drive an open discussion about recent advances in current and near-term commercially available genomic technologies. These technologies will include but are not limited to; single cell analysis, digital PCR, next generation sequencing, gene expression analysis. We have a panel of skilled scientists, each having common and unique expertise with numerous genome technologies. Join us to discuss best practices and share knowledge in this exciting field. Bring your questions, bring your challenges, bring your solutions, bring your expertise.
Track – Flow Cytometry
Title: Translational Medicine in Flow Cytometry
Location: Freedom Ballroom F
Speakers
- Erica Carpenter, PhD
Research Assistant Professor
Director of Circulating Tumor Material Center
University of Pennsylvania
- Beatriz Carreno, PhD
Research Associate Professor
University of Pennsylvania
Abstract
Flow cytometry enables the rapid analysis of heterogeneous cell populations, and provides multi-parameter information at the single-cell level. This session will highlight the critical role of flow cytometry in translational medicine.
Track – Imaging
Title: Image Analysis: Deconvolution “Seeing” through the Blur
Location: Freedom Ballroom E
Sponsor: Fluidigm
Speaker
- Richard Cole, PhD
State University of New York - Hosted by: Elke Küster-Schöck
Platform for Imaging by Microscopy
CHU Sainte-Justine/Université de Montréal, Montréal
Québec, Canada
Abstract
Any optical image system, such as a microscope, collects images which are a convolution of the “true” image, the distortion of the imaging system, plus noise. This is a characterizable modulation of the image and is defined as the Point Spread Function (PSF) of the optical system.
Image restoration by deconvolution is a mathematical process. The algorithm iteratively processes the “collected” image and returns a closer estimate of the “real” image based on a PSF and noise estimate. Whether the PSF is measured, estimated or theoretical, it can be used to correct the convoluted image. Since it’s possible to correct such a systematic error, we always should! When used with proper controls and deconvolution parameters the resulting image is quantifiable.
Track – Administration
Title: Core Administration Best Practices: Tools to Resolve Challenges in Core Administration (Business Plans, Advisory Boards, and Costing)
Location: Freedom Ballroom H
Speakers
- Mark Drinker MS
Assistant Director, Shared Resources
The Wistar Institute – Wistar Cancer Center
- Matthew Huesser, MBA
Enterprise Associate Director for Cancer Research Administration & Operations
Sidney Kimmel Cancer Center at Jefferson
- Jiju Mathew MBA
Manager – Financial Services/Service Centers
Department of Finance
University of Pennsylvania – Perelman School of Medicine
- Andrew Vinard (Host)
Director, Centralized Core Facilities
UMass Amherst, Institute for Applied Life Sciences
Abstract
An overview of tools in the administrator’s arsenal, including Advisory Boards, and Business Plans, and costing models. The session intends to review existing tools from the panelist’s institutions, and then be an open discussion on resources available at other institutions.
Keynote
Mark E. Curran, PhD
Vice President for Systems Pharmacology and Biomarkers
Janssen Research and Development
Title: Challenges in Pharmaceutical Development: Incorporating Big Data and Personalized Medicine
Abstract:
Despite advances in treatment of severe autoimmune diseases such as psoriasis, rheumatoid arthritis and inflammatory bowel disease, there remains a significant unmet clinical need for new therapies and integrated treatment solutions. At Janssen Immunology we are focused on transforming treatment of autoimmune diseases using state of the art molecular and cellular research, improved understanding of disease etiology, novel compound development platforms and precision medicine approaches. Our mission is to achieve higher response rates, deeper remission, interception and eventually prevention of these diseases. In this presentation I will describe progress and learning toward these objectives and how we are using systems pharmacology and data sciences to develop new medicines and improve the lives of patients living with autoimmune disease.
Speaker Bio
Mark Curran, PhD is Vice President, Head of Data Science for the Immunology Therapeutic Area at Janssen Research & Development, LLC. In this role, Mark is responsible for building an analytical group that will mine for and provide data that can inform and drive clinical trial design and pipeline decision-making. Mark’s work will generate and synthesize data across areas such as target identification, target validation, engineering of therapeutic molecules, and identification of selection and response biomarkers.
Mark joined Janssen in 2008, and was most recently Vice President, Systems Pharmacology and Biomarkers, where he was responsible for incorporating emerging science and human genetics into new programs to create more personalized treatment approaches predictive of response.
Before joining Janssen, Mark was Director of Biomarker Biology at Bristol-Myers Squibb where he worked across multiple therapeutic areas and led Early Development Teams conducting phase 1, 2 and experimental medicine trials for numerous development assets. Prior to that, Mark had roles of increased responsibility at DNA Sciences, ICAgen and Sequana Therapeutics/Axys Pharmaceuticals.
Mark earned a PhD in Human Genetics from the University of Utah and his undergraduate and masters’ degrees from Worcester Polytechnic Institute.
Location: Liberty Ballroom D
Time: 3:45 pm
Platinum Plus Vendor Sponsor Plenary – New England BioLabs, Inc.
Speaker
- Fiona Stewart
New England Biolabs, Inc.
Title: New Options for Methylome Analysis and RNA Depletion
Abstract
As the range of samples sequenced expands, the demands on NGS sample preparation increase, and improved options are required for applications including methylome analysis and transcriptome analysis.
Accurate identification of 5mC and 5hmC in DNA increases insight into potential gene regulatory mechanisms. Bisulfite sequencing is traditionally used to detect this methylation, but the chemical bisulfite reaction damages DNA, and results in libraries with high GC bias that are enriched for methylated regions. NEBNext Enzymatic Methyl-seq (EM-seq™) is an enzyme-based method for methylome analysis that minimizes DNA damage. EM-seq libraries have multiple performance advantages compared to bisulfite-converted libraries, including enabling more sensitive detection of 5mC and 5hmC, greater mapping efficiency and minimal GC bias.
In whole-transcriptome sequencing, highly expressed transcripts with minimal biological interest can dominate readouts, masking detection of more informative lower abundance transcripts. For applications where the full spectrum of RNA is of interest, or where poly(A) tails are absent or damaged, a method for depletion of abundant RNAs is required. Probe sets for the NEBNext RNaseH-based depletion method are being expanded to remove adult, fetal and embryonic globin transcripts from blood samples, to remove rRNA from bacteria, and to include a custom probe design tool.
Location: Liberty Ballroom D
Time: 4:45 pm
Poster Session & Vendor Show
Join us in the ballroom for light refreshments and to meet with our generous corporate sponsors, as well as visit the poster presenters.
Location: Liberty Ballroom D
Time: 5:00-6:30 pm
Platinum Plus Vendor Sponsor Plenary – Illumina
Speaker
- Jimmy Ortins
Executive Sales Specialist, Informatics
Illumina, Inc.
Title: Pairing the DRAGEN™ with the NovaSeq™ 6000 to Overcome Data Analysis Challenges in your Core
Abstract
Next Generation Sequencing (NGS) is an exciting, dynamic, and above all highly enabling technology for the study of genomics. New advances in NGS, such as the NovaSeq 6000 system from Illumina, are rapidly accelerating the rate at which genomics data is produced and can create some unique challenges in analyzing, storing, and sharing extremely large and valuable datasets. Today we will discuss the Illumina DRAGEN Bio-IT platform and highlight some of the unique characteristics that make it an especially valuable tool for Genomics and DNA sequencing core labs such as yours.
Location: Liberty Ballroom D
Time: 8:45 am
Keynote
Carl June, MD
University of Pennsylvania
Aaron Rapoport, MD
University of Maryland
Title: Development of Cell Therapies for Leukemia and Myeloma
Abstract
In recent years, immunotherapy has emerged as a promising treatment for certain types of cancer. A patient’s own immune cells, or T cells, can be genetically engineered to recognize and attack a cancer. Last year, the U.S. Food and Drug Administration (FDA) approved two versions of immunotherapy, which are known as chimeric antigen receptor (CAR) T-cell therapies – one for children and young adults with leukemia and another for adults with non-Hodgkin lymphoma.
Join us Friday morning, November 8, 2019, for this special keynote presentation as long-time collaborators and friends, Aaron P. Rapoport and Carl June, share the progress being made on T cell and vaccine immunotherapy.
Together they show how they and other physician scientists worldwide are carefully moving forward with T cell approaches, testing cancer vaccines and antibody-based therapies to stimulate the immune system to kill other kinds of cancer cells.
Speaker Bios
- Carl June, M.D., is a physician scientist and the Richard W. Vague Professor in Immunotherapy at the Perelman School of Medicine at the University of Pennsylvania in the Departments of Medicine and Pathology and Laboratory Medicine. He is the director of the Center for Cellular Immunotherapies at the Perelman School of Medicine, and the Director of the Parker Institute for Cancer Immunotherapy at the University of Pennsylvania. June has published more than 500 manuscripts and is the recipient of numerous prizes and honors, including election to the Institute of Medicine in 2012 and the American Academy of Arts and Sciences in 2014. He is the scientific founder of Xcyte Therapies Inc. and Tmunity Therapeutics. CTL019, the CAR T cell developed in the June laboratory was the first gene therapy to be approved by the US FDA in August 2017. The therapy developed by Dr. June’s team is now approved and marketed by Novartis in the US, Europe and Japan.
- Aaron P. Rapoport, M.D., is the Gary Jobson Professor of Medical Oncology and the Director of the Blood and Marrow Stem Cell Transplant and Cellular Therapy Programs at the Marlene and Stewart Greenebaum Comprehensive Cancer Center of the University of Maryland School of Medicine and Medical Center.
Dr. Rapoport received his bachelor’s degree in biology from Massachusetts Institute of Technology in 1982, and his M.D. from Harvard Medical School in 1986. He completed his medical internship and residency in internal medicine at Strong Memorial Hospital at the University of Rochester School of Medicine (1986-1989), and went on to both a clinical and research fellowship in hematology at the same institution (1989-1993).
Dr. Rapoport is a board-certified hematologist and stem cell transplant (SCT) physician with more than 30 years of experience in the care of patients with hematopoietic neoplasms and benign hematologic disorders. He also conducts translational research that focuses on the use of genetically modified autologous T cells and vaccines to enhance immune recovery and treat blood cancers including myeloma, lymphoma and leukemia. Dr. Rapoport was among the first to show that combination immunotherapy with vaccine-primed and ex-vivo costimulated autologous T-cells could induce robust vaccine-specific immune responses early after autologous stem cell transplantation (ASCT). He also demonstrated for the first time that early adoptive transfers of costimulated T cells could induce robust T-cell recovery post-transplant and elicit graft-versus-host disease-like reactions in the autologous transplant setting. In partnership with Dr. Carl June he helped to pioneer the use of genetically engineered T cells to treat multiple myeloma. He has directed six major clinical trials of adoptive T-cell therapy that have enrolled more than 150 patients. The clinical trials conducted by Dr. Rapoport and colleagues have provided a new avenue for rapid reconstitution of T-cell-mediated immunity following blood and marrow transplantation and laid the groundwork for the development of effective cellular immunotherapy of myeloma.
Over the course of his career, Dr. Rapoport has authored or co-authored more than 100 peer-reviewed journal articles. In 2016 Dr. Rapoport received the Clinical Research Achievement Award from the Clinical Research Forum for his paper entitled “ NY-ESO-1-Specific TCR-Engineered T cells Mediate Sustained Antigen-Specific Antitumor Effects in Myeloma” which was published in Nature Medicine. His work has been funded by the NIH, the Leukemia and Lymphoma Society (Clinical Scholars Award) and the Multiple Myeloma Research Foundation. He received the Stohlman Scholar award from the Leukemia and Lymphoma Society in 2006. In 2012, Dr. Rapoport was named the inaugural Gary Jobson Professor of Medical Oncology at the University of Maryland School of Medicine. He was also named a “Top Doc” by Baltimore Magazine from 2011-2019
Location: Liberty Ballroom D
Time: 9:00 am
Platinum Plus Vendor Sponsor Plenary – Agilent
Speaker
- Ji Zhu, Ph.D.
Senior Global Product Manager
Title: Magnis NGS Library Prep System, a Flexible and Fully Automated Solution for Your Lab
Abstract
One of the challenges with implementing NGS in a molecular laboratory is the expertise required to develop, optimize, and validate assays protocols for use in diverse applications. The Magnis system is comprised of an instrument, reagents, and protocols, and was designed to run complex NGS library assay with minimal technical knowledge and hands-on time.
Location: Liberty Ballroom D
Time: 10:00 am
Concurrent Sessions Block 4
Time: 10:45 am – 12:00 pm
Track – Genomics & Flow Cytometry
Title: Integrating Genomics and Flow: DNA+RNA+Protein = Integrated Molecular Diagnostics
Location: Freedom Ballroom G
Sponsor: Roche
Speaker
Toumy Guettouche
Director, Reagent Development, Roche Sequencing Solutions
Abstract
Roche’s New Target Enrichment Portfolio -Performance of Exome and Custom Panels including data from Short Hybridization and FFPE sample
Speakers
- Don A. Baldwin, PhD (Host)
Assoc. Professor of Pathology
Fox Chase Cancer Center
- Reza Nejati, MD
Assistant Professor of Pathology
Director of Flow Cytometry
Fox Chase Cancer Center
- Joseph M Breier, PhD
Digital Spatial Profiling-Technical sales Specialist
NanoString Technologies, Inc.
- Nina Luning Prak,, MD, PhD
Director, Human Immunology Core facility
Chair, Adaptive Immune Receptor Repertoire Community
Professor, Department of Pathology and Laboratory Medicine
Perelman School of Medicine. University of Pennsylvania
Abstract
This session will describe research and clinical applications that combine molecular phenotyping and genomics technologies. Multi-marker flow cytometry and highly multiplexed in situ spatial profiling can now be combined with DNA and RNA profiling to identify molecular mechanisms that are specific to precisely defined cell populations.
Recent cases of leukemia or lymphoma will be presented to illustrate how immunophenotyping, DNA and RNA profiling, and other molecular parameters are used for diagnosis and patient treatment. We will discuss current challenges in hematopathology that would benefit from translational technology development, particularly assessments for minimal residual disease and detection of tumor recurrence.
Track – Mass Spectrometry
Title: Mass Spectrometry: Rapid Tissue Imaging, Protein Digestion, Cysteine Adducts Identification
Location: Freedom Ballroom F
Speakers
- Robert Cole (Host)
Director, Mass Spectrometry and Proteomics Facility
Assistant Professor of Biological Chemistry
Johns Hopkins University
- Kristine Glunde, PhD
Radiology
Johns Hopkins University
- Raghothama Chaerkady, PhD
Dynamic Omics, Antibody Discovery and Protein Engineering
AstraZeneca
- Joshua Smith, PhD
Environmental Health
Johns Hopkins University
Abstract
Matrix-assisted Laser Desorption Ionization (MALDI) mass spectrometry imaging, which is also referred to as molecular histology, allows for histology-integrated visualization of metabolites, lipids, proteins, tryptic peptides, glycans, drugs, drug metabolites, and contrast agents. With new ion source technology and faster, more efficient lasers that are now commercially available, MALDI imaging has become a high-throughput, multiplexed tissue imaging approach that can spatially map up to 5,000 biomolecules at once in clinically relevant times, at spatial resolutions of 20 μm pixel size or better. The ease of use of commercial MALDI mass spectrometers that are set up for imaging has also dramatically increased, and robotic sprayers that allow for reproducible matrix deposition have become commercially available, making this technology now ready for the core facility setting. This presentation will introduce the technology, detail the MALDI imaging workflow including tips and tricks of sample preparation, and showcase several examples of the application of MALDI imaging in biomedical research studies.
Track – Imaging
Title: Microscopy: Enhancing Reproducibility of Microscopy Data Through Quality Control and Data Management
Location: Freedom Ballroom E
Speakers
- Michelle Itano, Ph.D.
University of North Carolina Chapel Hill
- Caterina Strambio-De-Castillia, Ph.D.
University of Massachusetts
- Jason Swedlow, Ph.D.
University of Dundee
- Jay Copeland, M.S.
Harvard University
Abstract
Currently, researchers are forced to manually track and annotate all imaging experiment steps, from sample preparation, to image data acquisition, storage, processing, and analysis, which is very onerous, slow, and wasteful. Lack of automation and poor documentation often makes it impossible to reproduce, compare, and combine large sets of data from different laboratories, biological systems, or different experimental setups, making it difficult for scientists to collaborate with each other. This session will address these issues by presenting recent tools and advances to enhance image quality control and data management.
Track – Administration & Bioinformatics
Title: Data Management and Analysis Cores and Making Research FAIR Compliant
Location: Freedom Ballroom H
Speakers
- Stuart Levine, Ph.D.
Director MIT BioMicro Center
Massachusetts Institute of Technology
- Cathy Wu, PhD.
Unidel Edward G. Jefferson Chair in Engineering and Computer Science
Professor, Departments of Computer & Information Sciences and Biological Sciences
University of Delaware
Abstract
This session will focus on practical measures in creating and maintaining Data Management and Analysis Cores as well as keys in ensuring data maintains FAIR compliance (making data Findable, Accessible, Interoperable and Reproducible. The session will be well suited for Bioiformaticists as well as Administrators and will be of interest to any lab creating ‘omic scale data.
Closing Remarks
Speakers
- Stuart Levine
NERLSCD President
- Kevin Gerrish
MADSSCI President
Time: 12:00-1:00 pm
Location: Liberty Ballrooms C&D
Core Facilities tours
All tours will meet in the Hotel 201 lobby. The bus will depart at 1:00 pm
- Tour 1:
Cellular Immunotherapies Cores, Smilow Research Building, Penn Clinical Manufacturing Suite including Translational and Correlative Lab supporting Clinical Trials
Tour 2:
Wistar and Penn Cores (Groups will ‘swap’ at 2:00—to see both institutions’ cores)
Penn: Flow, Electron Microscopy, DNA Sequencing and Human Immunology
Wistar: Flow, Genomics and Imaging
Poster Abstracts
- bioMT Institute for Biomolecular targeting
- Title: bioMT: Molecular Tools Core, a Protein Production Wizard
- Authors: Verissimo A, Kull A, Svindrych Z, Shipman E & Madden D
- Affiliations: bioMT, Institute for Biomolecular Targeting, Geisel School of Medicine at Dartmouth, Hanover, NH United States
- Abstract: Selecting a method to produce a recombinant protein is not an easy task and there is no such thing as “one-size fits all “. Often researchers face a bewildering array of choices such as which host, vector or strains to choose. They need to decide if expressing the full-length protein or just a fragment and which affinity tag is best for each individual protein and suitable for the intended downstream application. Lastly, an appropriate purification strategy must be developed and polished to produce high quality recombinant proteins. Unfortunately, because every protein is different there are no standard protocols and expression and purification must be worked out in each case. The bioMT Molecular Tools Core enables research projects by assisting researchers on the decision-making process, on the optimization of expression conditions and development of purification schemes, providing the necessary training and expertise along the way. In addition, the MTC also executes and develops independently new protocols to produce high-quality recombinant proteins. Currently, we are developing a robust pipeline for protein production in bacterial, mammalian and insect cells and will include yeast expression systems in our toolbox soon. All proteins are produced under standardized conditions to ensure that consistent, reproducible and reliable products. Here we show a few examples of how the MTC has provided support to researchers and contributed to the advancement of their research projects. The bioMT looks forward to expanding its user base and becoming a pivotal resource to all Dartmouth community.
- University of Pennsylvania
- Title: Evaluation of short-read NGS methods for detection of gene editing effects in translational research for gene therapy.
- Authors: Copren, K; Saveliev, A; Latshaw, C; Liu, J; Burashnikov, E; Yan, H; Sims, J; Lee, C; Wilson, JM.
- Affiliations: Nucleic Acid Technologies Core, Gene Therapy Program, Perelman School of Medicine, University of Pennsylvania
- Abstract: Genome editing is a powerful tool in disease research. In 2019 the first clinical trials using genome editing in monogenic diseases were approved, and the field is advancing rapidly. However, detecting both the off- and on-target effects of the nucleases pose challenges for assay design. Sensitive and accurate detection of these effects is imperative. The GTP NAT-Core has optimized and regularly runs a variety of short read NGS pipelines specifically to detect gene editing effects including a general targeted amplicon sequencing method, Guide-Seq, and AMP-Seq. We will compare and contrast the sensitivity, accuracy, throughput and turnaround time of each of these methods for the complete lab to bioinformatic pipeline—all variables that require thoughtful validation to meet the demanding rigor of advancing these technologies through translational research and into clinical trials. We will discuss continuing strategies and development challenges using short-read NGS for the accurate measurement of gene editing success.
- Columbia University
- Title: Quantitative Proteomics and Metabolomics Center
- Author: Brown, Lewis M.
- Affiliations: Columbia University, New York, NY
- Abstract: We do discovery, targeted, proximity labeling or posttranslational modification studies of patient plasma, CSF, tissues (including FFPE), cultured cells, affinity/IP pulldowns, organelles, yeast, Drosophila, E. coli, and many other sample types. We measure protein abundance not just ratios. We support collaborative proposal development with rapid response for research plans, experimental & statistical design, preliminary data, resource & equipment descriptions, budgets, biosketches and responses to study sections. In an example project presented today, we show that mesenchymal stem cells (MSCs) can suppress inflammation and promote tissue repair. Hypoxia and inflammation are often present in acute injury and chronic disease, so we sought to explore how the proteome of MSCs changes when cells were exposed to hypoxia and/or Interferon-gamma (IFN-gamma) treatment individually or in combination using label-free protein profiling as well as metabolite profiling.
- Nemours A.I.duPont Hospital for Children
- Title: Team Based Implementation of a Pharmacogenomics Pipeline at Nemours Children’s Health System
- Authors: Stabley D1, Holbrook J1, Vinette K1, Hirata L1, Pannah N1, Cook KJ2,3, Duong BQ1, Kirwin S1, Robbins K1, Funanage VL1, Blake K2
- Affiliations: 1Nemours/Alfred I duPont Hospital for Children, Wilmington, DE. 2Nemours Children’s Specialty Care, Jacksonville, FL. 3University of Florida College of Pharmacy, Jacksonville, FL
- Abstract: Each year the Nemours Children’s Health System (NCHS), a nonprofit, multi-state pediatric health system, provides care to 410,000 children in the Delaware Valley and Florida. In 2018, out of 277,581 NCHS patients who received a drug, 51,791 patients (18.6%) received at least one drug with a significant drug-gene interaction according to the Clinical Pharmacogenetics Implementation Consortium (CPIC). Within that same year and population, out of the 51,791 patients that received at least one drug with significant drug-gene interaction, 11,187 patients (21.6%) received two or more drugs with PGx interactions. Due to the significance of PGx implications, NCHS implemented the Clinical Pharmacogenomics Service under the Nemours Precision Medicine program. Here we explain the collaboration between the Precision Medicine program, Molecular Diagnostic Laboratory, and Biomolecular Core Laboratory and the development of the Nemours in-house PGx panel. Utilizing the ThermoFisher TaqMan® Assay Custom 384 well plating format and the QuantStudio™ 12K FLEX platform, we designed a customized PGx panel that contains 48 genotyping assays. This plate design includes a mix of both inventoried and custom designed genotyping assays. Additionally, three CYP2D6 copy number assays were included in the PGx pipeline because copy number also impacts CYP2D6 phenotype. After analysis with Genotyper™ and AlleleTyper™, results are uploaded as discrete data into the electronic health record (EHR). We will discuss how to troubleshoot genotyping and copy number issues that arise using different specimen types, i.e. saliva versus blood. Finally, we will discuss how to develop a pipeline for both the validation and the analysis of the genotyping and copy number PGx data.
- Massachusetts Institute of Technology
- Title: Miniaturization of Illumina Library Preparation using Robotic Low-volume Liquid Handler
- Authors: Mildrum S1, Hendricks A1, Kamelamela N1, Levine S1
- Affiliations: . MIT BioMicro Center, Massachusetts Institute of Technology, Cambridge, MA, USA
- Abstract: Due to the high cost of Illumina library production, many NGS experiments have succumbed to the utilization of underpowered experiments; however, high-throughput library preparation offers a unique alternative to circumvent the high experimental costs associated with normal library preparation. To address the cost of library preparation, we have optimized a diverse set of Illumina library preparation to work with minimal reagents using the TTP Labtech Mosquito HV. Here we demonstrate several of our miniaturized protocols and their application to a broad variety of biological sample types. These methods for both RNA and DNA library production from either intact or fragmented samples have been successfully implemented and are widely used. Consequently, the miniaturization of these protocols has resulted in significant savings in library production costs on a per sample basis, which enables the scientist to increase the number of replicates per condition and ultimately to enhance the power of their NGS experiment without increasing the total amount of sequencing required.
- Albert Einstein College of Medicine
- Title: EPIGENOMICS SHARED FACILITY
- Authors: MAQBOOL, S.B., OLEA, R., MI, S., ZHAO, Y., AND GREALLY, J.M
- Affiliations: Albert Einstein College of Medicine, Bronx, NY
- Abstract: The Epigenomics Shared Facility (ESF), part of Einstein’s Center for Epigenomics and an Illumina CSPro (certified service provider) laboratory, offers massively-parallel sequencing (MPS) including fully-automated library preparation, quality control and assurance, and a number of assays to study the epigenome. The high-throughput molecular technological resources in the ESF include massively parallel sequencing (MPS) platforms Illumina HiSeq2500, Illumina NEXTSeq500, Illumina MiSeq, Roche FLX; and TECAN freedom Evo® 200 Robotics. Sample information is uploaded through a web-based interface (WASP) prior to sample submission, allowing LIMS and automated primary analysis of the data generated using the Wasp System software, and returning data through visualization and web links. Data analytical services are provided by the Computational and Statistical Epigenomics Group. We encourage novel research in a number of human diseases, with early emphases on cancer, neuroepigenomics, the epigenomics of infectious disease, aging research, diabetes and renal disorders
- MIT
- Title: SYBR and A260/A280 Quantification of DNA as additional tools in High Throughput Library Setups
- Authors: Vishwanath V1, 2, Morgan C1, 2, Ahmed S1, Kim T1, Levine S1, 2
- Affiliations: 1: Massachusetts Institute of Technology, BioMicro Center, Department of Biology, 31 Ames Street, Cambridge, MA 02140; 2: Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, 400 Main Street, Cambridge, MA 02140
- Abstract: As single cell transcriptome sequencing continues to expand and evolve, high throughput setups to NGS library preparation need to overcome several constraints, particularly those with time, sample integrity, and costs. Here, we present SYBR quantification via the VarioSkan plate reader as well as A260/A280 measurements as viable options to consistently quantify libraries in a fashion that is also compatible with automation. Using sequencing analysis data, we show that the values obtained through these quantification methods can serve as fairly accurate guidelines for multiplexing and downstream sequencing and analysis, and can reduce the need for NanoDrop quantitation and/or qPCR for individual samples in high-throughput setups.
- Zymo Research Corp
- Title: Examination of two leading commercial kits for rRNA depletion and poly(A)-enrichment and a novel probe-free method to compare depleted/enriched samples with untreated samples.
- Author: Justin Lin
- Affiliations: Zymo Research Corp
- Abstract: RNA-Seq is a powerful tool that allows researchers to effectively digitize the transcriptome of a biological sample. However, ribosomal RNA (rRNA) transcripts typically comprise between 80%-90% of all cellular RNAs and must be avoided or eliminated for researchers to efficiently analyze the remaining 10% of biologically relevant sequences. Oligo(T)-mediated mRNA-enrichment and probe-based ribosomal depletion are the two of the most common methods used to avoid or eliminate rRNA reads from RNA-seq libraries. In the former, only mRNA transcripts with sufficiently long poly-adenylated tails are captured, and in the latter, high concentrations DNA probe-sets are used to capture and eliminate complementary rRNA transcripts. However, no study to date has provided a solution to address the effects of off-target bias in either ribosomal depletion or poly(A)-enrichment on the whole transcriptome. Here, we examine two leading commercial kits for rRNA depletion and poly(A)-enrichment and a novel probe-free method to compare depleted/enriched samples with untreated samples. Our comparison reveals that conventional, probe-based ribosomal depletion and oligo(T)-mediated poly(A)-enrichment methods both introduce significant changes to the transcriptome, while the probe-free method is effectively free of bias and best preserves the original transcriptome profile. We find that the probe-free method is universally applicable to any sample type, even across samples representing organisms from different biological kingdoms and phyla.
- National Institutes of Health (Leidos Biomedical)
- Title: Impact of fecal microbiome extraction technique on relative abundance of genera within expected and unexpected communities
- Authors: Sevilla S1,2, Vogtmann E1, Abnet C1, Sinha R1, Hicks B1,2, Jones K1,2, Hutchinson A1,2, Dagnall C1,2
- Affiliations: 1 Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI), National Institutes of Health (NIH), Bethesda, MD, USA 2 Cancer Genomics Research Laboratory, Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research, Frederick, MD, USA
- Abstract: Impact of fecal microbiome extraction technique on relative abundance of genera within expected and unexpected communities. Microbiome analysis has gained significant interest as sequencing technology has improved, allowing for the examination of microbial communities often believed to play a key role in human health and disease. 16S rRNA sequencing analysis, utilizing various publicly available bioinformatic tools, allows the study of diversity in microbial communities. It has previously been shown that extraction techniques are impactful for the analysis, however, these studies have often been performed with a single cultured sample, or with evenly distributed mock communities, failing to provide variability often seen with human source samples and excluding the impact of amplification and/or sequencing related bias. Within this study, we analyzed eight different extraction protocols, utilizing commercially available reagents, with a fresh-frozen human fecal sample, and mock communities, of varied evenness and species distribution. In addition, six sequencing controls, variable concentration and species distribution, were utilized to determine the additional bias introduced by amplification, sequencing reagents and bioinformatic parameters. Significant differences were noted in alpha and beta diversity metrics with each protocol, as well as to the expected values. Additionally, unexpected genera were found within each protocol indicating uniquely introduced contamination. Trends were noted between different kits, in over- or under-representation of specific genera of communities. Similarly, trends were observed sequencing controls, indicating the contamination and/or bias introduced after extraction. Overall, these findings illustrate the importance of utilizing controls at multiple stages of the process, to better assess the impact of the findings of any human samples.”
| Location | Vendor | Information |
| 28 | ABRF | |
| 17 | Advaita Bioinformatics | Next Generation Bioinformatics |
| 21 | Agilent | Assorted Lab Products, instruments, chemical analysis and diagnostic |
| no booth | BioLegend | Antibiotics and biomedical research |
| 32 | BMG LabTech | Microplate reader instrumentation |
| 29 | CORIS Life and Sciences Monitoring | Temperature monitoring and alerts |
| 7 | Covaris Inc | Mass spetrometry and proteomic analysis (SMAP) |
| 33 | Cytek Biosciences, Inc | Flow Cytometry |
| 4 | De Novo Software | Data analysis, academic, clinic, validations, analysis cytometry data |
| 30 | Eclipse Bio | RNA Genomics Solutions |
| 6 | Elsevier/Cell | Publishing |
| 31 | Eppendorf NA | Lab equipment and lab consumables |
| 5 | Fluidigm | fluidics Mass Cytometry |
| 18 | GE Healthcare Lifesciences | NGS prep, magnetic beads, Genomics kits, cell culture, super-res, high-res, and HCA microscopy, chromatography, protein purification, detection, and imaging analysis, biomolecular interaction analysis, sample filtration, upstream and downstream bioprocess, cell and gene therapy |
| 20 | Illumina | NGS, Microarray scanners, In Vitro diagnosics (IVD) |
| 19 | Integra Biosciences | Pipettes, pumps, media prep |
| 14 | Integrated DNA Technologies | Oligos, CRISPR genome editing, Reagents and kits |
| 39 | Lexogen | Kits RNA extraction from blood, Total RNA-Seq library kit, Total cDNA amplification kit |
| 12 | Luminex Corporation | ELISA, clinical |
| 11 | Macherey-Nagel | Products for filtration, water analysis, chromoatography, rapid test strips – urine |
| 13 | NanoCellect Biomedical Inc | Cell Sorter, microfluidics |
| 38 | New Englan Biolabs Inc | Supplies recombinant and native enzyme reagents Related to New England Biolabs and Illumina – NEB, Agilent, Qiagen, Thermofisher, Illumina |
| 10 | Oxford Nanopore Technolgies Ltd | Nanopore sequencing, NGZ |
| 37 | Partek | Microarry, NGS, DNA analysis |
| 36 | PerkinElmer | Instruments, Fluorescent probes, clinical analysis |
| 9 | Promega Corporation | Enzymes, Nucleic acid kits, fluorometers |
| 34 | Qiagen | NGS Kits, RNA sequencing kits, sample prep |
| 15 | Refeyn Ltd | Mass Photometry- sample purity, mass measurement, protein – protein interactions |
| 16 | Roche Sequencing and Life Sciences | Oncology assays, library prep, hybrid-capture target enrichment |
| 8 | SeqWell | NGS library preparation |
| 35 | StemCell Tech | Cell Culture media and cells |
| 1 | Swift Biosciences | DNA, RNA library kits |
| 2 | Takara Bio USA inc | NGS, Gene funcction, stem cell researh, PCR, cloning, Ab and Elisa |
| 22 | Tecan Genomics | NA purtification, RNA and DNA quantification, PCR setup, CRISP-Cas9 Gemome editing |
| 26 | ThermoFisher (Flow) | Flow Cytometry |
| 3 | TTP LabTech | Drug discovery, liguid handling equipment, DNA storage -80°C |
| 23 | Twist Bioscience | DNA synthesis for disease related disease, and therapeutics |
| 25 | Zymo Research Corporation | Total RNA library kits, DNA kits |